Co-Authored with Jared Dunnmon (Stanford University)
Medical AI testing is unsafe, and that isn’t likely to change anytime soon.
No regulator is seriously considering implementing “pharmaceutical style” clinical trials for AI prior to marketing approval, and evidence strongly suggests that pre-clinical testing of medical AI systems is not enough to ensure that they are safe to use. As discussed in a previous post, factors ranging from the laboratory effect to automation bias can contribute to substantial disconnects between pre-clinical performance of AI systems and downstream medical outcomes. As a result, we urgently need mechanisms to detect and mitigate the dangers that under-tested medical AI systems may pose in the clinic.
In a recent preprint co-authored with Jared Dunnmon from Chris Ré’s group at Stanford, we offer a new explanation for the discrepancy between pre-clinical testing and downstream outcomes: hidden stratification. Before explaining what this means, we want to set the scene by saying that this effect appears to be pervasive, underappreciated, and could lead to serious patient harm even in AI systems that have been approved by regulators.
But there is an upside here as well. Looking at the failures of pre-clinical testing through the lens of hidden stratification may offer us a way to make regulation more effective, without overturning the entire system and without dramatically increasing the compliance burden on developers.
What’s in a stratum?
We recently published a pre-print titled “Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging“.
Note: While this post discusses a few parts of this paper, it is more intended to explore the implications. If you want to read more about the effect and our experiments, please read the paper 🙂
The effect we describe in this work — hidden stratification — is not really a surprise to anyone. Simply put, there are subsets within any medical task that are visually and clinically distinct. Pneumonia, for instance, can be typical or atypical. A lung tumour can be solid or subsolid. Fractures can be simple or compound. Such variations within a single diagnostic category are often visually distinct on imaging, and have fundamentally different implications for patient care.

Examples of different lung nodules, ranging from solid (a), solid with a halo (b), and subsolid (c). Not only do these nodules look different, they reflect different diseases with different patient outcomes.
We also recognise purely visual variants. A pleural effusion looks different if the patient is standing up or is lying down, despite the pathology and clinical outcomes being the same.
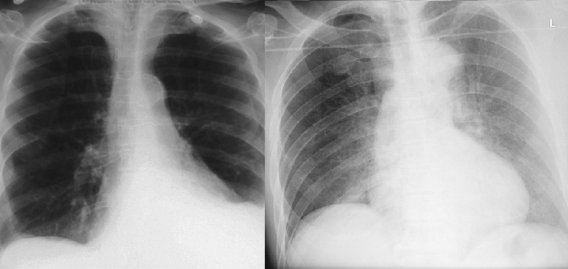
These patients both have left sided pleural effusions (seen on the right of each image). The patient on the left has increased density at the left lung base, whereas the patient on the right has a subtle “veil” across the entire left lung.
These visual variants can cause problems for human doctors, but we recognise their presence and try to account for them. This is rarely the case for AI systems though, as we usually train AI models on coarsely defined class labels and this variation is unacknowledged in training and testing; in other words, the the stratification is hidden (the term “hidden stratification” actually has its roots in genetics, describing the unrecognised variation within populations that complicates all genomic analyses).
The main point of our paper is that these visually distinct subsets can seriously distort the decision making of AI systems, potentially leading to a major difference between performance testing results and clinical utility.
Clinical safety isn’t about average performance
The most important concept underpinning this work is that being as good as a human on average is not a strong predictor of safety. What matters far more is specifically which cases the models get wrong.
For example, even cutting-edge deep learning systems make such systematic misjudgments as consistently classifying canines in the snow as wolves or men as computer programmers and women as homemakers. This “lack of common sense” effect is often treated as an expected outcome of data-driven learning, which is undesirable but ultimately acceptable in deployed models outside of medicine (though even then, these effects have caused major problems for sophisticated technology companies).
Whatever the risk is in the non-medical world, we argue that in healthcare this same phenomenon can have serious implications.
Take for example a situation where humans and an AI system are trying to diagnose cancer, and they show equivalent performance in a head-to-head “reader” study. Let’s assume this study was performed perfectly, with a large external dataset and a primary metric that was clinically motivated (perhaps the true positive rate in a screening scenario). This is the current level of evidence required for FDA approval, even for an autonomous system.
Now, for the sake of the argument, let’s assume the TPR of both decision makers is 95%. Our results to report to the FDA probably look like this:

TPR is the same thing as sensitivity/recall
That looks good, our primary measure (assuming a decent sample size) suggests that the AI and human are performing equivalently. The FDA should be pretty happyª.
Now, let’s also assume that the majority of cancer is fairly benign and small delays in treatment are inconsequential, but that there is a rare and visually distinct cancer subtype making up 5% of all disease that is aggressive and any delay in diagnosis leads to drastically shortened life expectancy.
There is a pithy bit of advice we often give trainee doctors: when you hear hoofbeats, think horses, not zebras. This means that you shouldn’t jump to diagnosing the rare subtype, when the common disease is much more likely. This is also exactly what machine learning models do – they consider prior probability and the presence of predictive features but, unless it has been explicitly incorporated into the model, they don’t consider the cost of their errors.
This can be a real problem in medical AI, because there is a less commonly shared addendum to this advice: if zebras were stone cold killing machines, you might want to exclude zebras first. The cost of misidentifying a dangerous zebra is much more than that of missing a gentle pony. No-one wants to get hoofed to death.

In practice, human doctors will be hyper-vigilant about the high-risk subtype, even though it is rare. They will have spent a disproportionate amount of time and effort learning to identify it, and will have a low threshold for diagnosing it (in this scenario, we might assume that the cost of overdiagnosis is minimal).
If we assume the cancer-detecting AI system was developed as is common practice, it probably was trained to detect “cancer” as a monolithic group. Since only 5% of the training samples included visual features of this subtype, and no-one has incorporated the expected clinical cost of misdiagnosis into the model, how do we expect it to perform in this important subset of cases?
Fairly obviously, it won’t be hypervigilant – it was never informed that it needed to be. Even worse, given the lower number of training examples in the minority subtype, it will probably underperform for this subset (since performance on a particular class or subset should increase with more training examples from that class). We might even expect that a human would get the majority of these cases right, and that the AI might get the majority wrong. In our paper, we show that existing AI models do indeed show concerning error rates on clinically important subsets despite encouraging aggregate performance metrics.
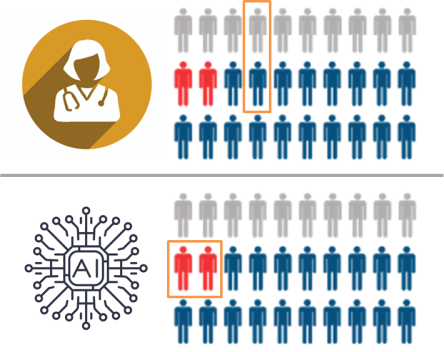
In this hypothetical, the human and the AI have the same average performance, but the AI specifically fails to recognise the critically important cases (marked in red). The human makes mistakes in less important cases, which is fairly typical in diagnostic practice.
In this setting, even though the doctors and the AI have the same overall performance (justifying regulatory approval), using the AI would lead to delayed diagnosis in the cases where such a delay is critically important. It would kill patients, and we would have no way to predict this with current testing.
Predicting where AI fails
So, how can we mitigate this risk? There are lots of clever computer scientists trying to make computers smart enough to avoid the problem (see: algorithmic robustness/fairness, causal machine learning, invariant learning etc.), but we don’t necessarily have to be this fancy^. If the problem is that performance may be worse in clinically important subsets, then all we might need to do is identify those subsets and test their performance.
In our example above, we can simply label all the “aggressive sub-type” cases in the cancer test set, and then evaluate model performance on that subset. Then our results (to report to the FDA would be):

As you might expect, these results would be treated very differently by a regulator, as this now looks like an absurdly unsafe AI system. This “stratified” testing tells us far more about the safety of this system than the overall or average performance for a medical task.
So, the low-tech solution is obvious – you identify all possible variants in the data and label them in the test set. In this way, a safe system is one that shows human-level performance in the overall task as well as in the subsets.
We call this approach schema completion. A schema (or ontology) in this context is the label structure, defining the relationships between superclasses (the large, coarse classes) and subclasses (the fine-grained subsets). We have actually seen well-formed schemas in medical AI research before, for example in the famous 2017 Nature paper Dermatologist-level classification of skin cancer with deep neural networks by Esteva et al. They produced a complex tree structure defining the class relationships, and even if this is not complete, it is certainly much better than pretending that all of the variation in skin lesions is explained by “malignant” and “not malignant” labels.

So why doesn’t everyone test on complete schema? Two reasons:
- There aren’t enough test cases (in this dermatology example, they only tested on the three red super-classes). If you had to have well-powered test sets for every subtype, you would need more data than in your training set!
- There are always more subclasses*. In the paper, Esteva et al describe over 2000 diagnostic categories in their dataset! Even then they didn’t include all of the important visual subsets in their schema, for example we have seen similar models fail when skin markers are present.
So testing all the subsets seems untenable. What can we do?
We think that we can rationalise the problem. If we knew what subsets are likely to be “underperformers”, and we use our medical knowledge to determine which subsets are high-risk, then we only need to test on the intersection between these two groups. We can predict the specific subsets where AI could clinically fail, and then only need to target these subsets for further analysis.
In our paper, we identified three main factors that appear to lead to underperformance. Across multiple datasets, we find evidence that hidden stratification leads to poor performance when there are subsets characterized by low subset prevalence, poor subset label quality, and/or subtle discriminative features (when the subset looks more like a different class than the class that it actually belongs to).

An example from the paper using the MURA dataset. Relabeled, we see that metalwork (left) is visually the most obvious finding (it looks the least like a normal x-ray out of the subclasses). Fractures (middle) can be subtle, and degenerative disease (right) is both subtle and inconsistently labeled. A model trained on the normal/abnormal superclasses significantly underperforms on cases within the subtle and noisy subclasses.
Putting it into practice
So we think we know how to recognise problematic subsets.
To actually operationalise this, we doctors would sit down and write out a complete schema for any and all medical AI tasks. Given the broad range of variation, covering clinical, pathological, and visual subsets, this would be a huge undertaking. Thankfully, it only needs to be done once (and updated rarely), and this is exactly the sort of work that is performed by large professional bodies (like ACR, ESR, RSNA), who regularly form working groups of domain specialists to tackle these kind of problems^^.

The nicest thing you can say about being in a working group is that someone’s gotta do it.
With these expert-defined schema, we would then highlight the subsets which may cause problems – those that are likely to underperform due to the factors we have identified in our research, and those that are high risk based on our clinical knowledge. Ideally there will be only a few “subsets of concern” per task that fulfil these criteria.

Then we present this ontology to the regulators and say “for an AI system to be judged safe for task X, we need to know the performance in the subsets of concern Y and Z.” In this way, a pneumothorax detector would need to show performance in cases without chest tubes, a fracture detector would need to be equal to humans for subtle fractures as well as obvious ones, and a “normal-case detector” (don’t get Luke started) would need to show that it doesn’t miss serious diseases.
To make this more clear, let’s consider a simple example. Here is a quick attempt at a pneumothorax schema:

Subsets of concern in red, conditional subsets of concern in orange (depends on exact implementation of model and data)
Pneumothorax is a tricky one since they are all “high risk” if they are untreated (meaning you end up with more subsets of concern than in many tasks), but we think this gives a general feel for what schema completion might look like.
The beauty of this approach is that it would work within the current regulatory framework, and as long as there aren’t too many subsets of concern the compliance cost should be low. If you already have enough cases for subset testing, then the only cost to the developer would be producing the labels, which would be relatively small.
If the subsets of concern in the existing test set are too small for valid performance results, then there is a clear path forward – you need to enrich for those subsets (i.e., not gather ten thousand more random cases). While this does carry a compliance cost, since you only need to do this for a small handful of subsets, the cost is also likely to be small compared to the overall cost of development. Sourcing the cases could get tricky if they are rare, but this is not insurmountable.
The only major cost to developers when implementing a system like this is if they find out that their algorithm is unsafe, and it needs to be retrained with specific subsets in mind. Since this is absolutely the entire point of regulation, we’d call this a reasonable cost of doing business.
In fact, since this list of subsets of concern would be widely available, developers could decide on their AI targets informed of the associated risks – if they don’t think they can adequately test for performance in a subset of concern, they can target a different medical task. This is giving developers have been asking for – they say they want more robust regulation and better assurances of safety, as long as the costs are transparent and the playing field is level.
How much would it help?
We see this “low-tech” approach to strengthen pre-clinical testing as a trade-off between being able to measure the actual clinical costs of using AI (as you would in a clinical trial) and the realities of device regulation. By identifying strata that are likely to produce worse clinical outcomes, we should be able to get closer to the safety profile delivered by gold standard clinical testing, without massively inflating costs or upending the current regulatory system.
This is certainly no panacea. There will always be subclasses and edge cases that we simply can’t test preclinically, perhaps because they aren’t recognised in our knowledge base or because examples of the strata aren’t present within our dataset. We also can’t assess the effects of the other causes of clinical underperformance, such as the laboratory effect and automation bias.
To close this safety gap, we still need to rely on post-deployment monitoring.
A promising direction for post-deployment monitoring is the AI audit, a process where human experts monitor the performance and particularly the errors of AI systems in clinical environments, in effect estimating the harm caused by AI in real-time. The need for this sort of monitoring has been recognised by professional organisations, who are grappling with the idea that we will need a new sort of specialist – a chief medical information officer who is skilled in AI monitoring and assessment – embedded in every practice (for example, see section 3 of the proposed RANZCR Standards of Practice for Artificial Intelligence).
Auditors are the real superheros
Audit works by having human experts review examples of AI predictions, and trying to piece together an explanation for the errors. This can be performed with image review alone or in combination with other interpretability techniques, but either way error auditing is critically dependent on the ability of the auditor to visually appreciate the differences in the distribution of model outputs. This approach is limited to the recognition of fairly large effects (i.e., effects that are noticeable in a modest/human-readable sample of images) and it will almost certainly be less exhaustive than prospectively assessing a complete schema defined by an expert panel. That being said, this process can still be extremely useful. In our paper, we show that human audit was able to detect hidden stratification that caused the performance of a CheXNet-reproduction model to drop by over 15% ROC-AUC on pneumothorax cases without chest drains — the subset that’s most important! — with respect to those that had already been treated with a chest drain.
Thankfully, the two testing approaches we’ve described are synergistic. Having a complete schema is useful for audit; instead of laboriously (and idiosyncratically) searching for meaning in sets of images, we can start our audit with the major subsets of concern. Discovering new and unexpected stratification would only occur when there are clusters of errors which do not conform to the existing schema, and these newly identified subsets of concern could be folded back into the schema via a reporting mechanism.
Looking to the future, we also suggest in our paper that we might be able to automate some of the audit process, or at least augment it with machine learning. We show that even simple k-means clustering in the model feature space can be effective in revealing important subsets in some tasks (but not others). We call this approach to subset discovery algorithmic measurement, and anticipate that further development of these ideas may be useful in supplementing schema completion and human audit. We have begun to explore more effective techniques for algorithmic measurement that may work better than k-means, but that is a topic for another day :).
Making AI safe(r)
These techniques alone won’t make medical AI safe, because they can’t replace all the benefits of proper clinical testing of AI. Risk-critical systems in particular need randomised control trials, and our demonstration of hidden stratification in common medical AI tasks only reinforces this point. The problem is that there is no path from here to there. It is possible that RCTs won’t even be considered until after we have a medical AI tragedy, and by then it will be too late.
In this context, we believe that pre-marketing targeted subset testing combined with post-deployment monitoring could serve as an important and effective stopgap for improving AI safety. It is low tech, achievable, and doesn’t create a huge compliance burden. It doesn’t ask the healthcare systems and governments of the world to overhaul their current processes, just to take a bit of advice on what specific questions need to be asked for any given medical task. By delivering a consensus schema to regulators on a platter, they might even use it.
And maybe this approach is more broadly attractive as well. AI is not human — inhuman, in fact — in how it makes decisions. While it is attractive to work towards human-like intelligence in our computer systems, it is impossible to predict if and when this might be feasible.
The takeaway here is that subset-based testing and monitoring is one way we can bring human knowledge and common sense into medical machine learning systems, completely separate from the mathematical guts of the models. We might even be able to make them safer without making them smarter, without teaching them to ask why, and without rebooting AI.


Dear Dr. Luke,
Your posts and insights regarding this new field are always out-of-this-world! I’ve been reading and following your work for more than 2 years and its quality never stops to amaze me! Many congratulations on this and keep up the unbelievably great work!
If I may, it would be great to have your feedback on this: our startup is using ml for sepsis early detection and I was wondering what would be the best approach to avoid the perils of hidden stratification? Using algorithmic measurement to come up with subsets and make sure our sensitivity is high for subsets with higher mortality rate (assuming there are subsets with distinct mortality rate)?
Thank you very much for your help!
Kindest regards, Goncalo, M.D. CEO
On Mon, Oct 14, 2019 at 12:26 PM Luke Oakden-Rayner wrote:
> lukeoakdenrayner posted: “Co-Authored with Jared Dunnmon (Stanford > University) Medical AI testing is unsafe, and that isn’t likely to change > anytime soon. No regulator is seriously considering implementing > “pharmaceutical style” clinical trials for AI prior to marketing approv” >
LikeLike
Thanks Goncalo!
If you have mortality rate as an explicit recorded variable, then the first thing I would do is visualise the relationship between sensitivity and mortality. Hopefully sensitivity does not have any dips in the high mortality region.
But otherwise I think all you can do is use your knowledge to define subsets of concern, like we propose. Since you aren’t using visual data, you will be looking at more traditional stratifying factors. Age, gender, comorbidities, biomarkers etc.
I guess try to make sure that subgroups of patients that are much smaller (ie children, people with rare but significant-to-the-task disorders like immunosuppression etc) don’t show vastly worse performance.
LikeLike
Nice article! However, the comment about FDA is a little misleading (this one: “Let’s assume this study was performed perfectly, with a large external dataset and a primary metric that was clinically motivated (perhaps the true positive rate in a screening scenario). This is the current level of evidence required for FDA approval, even for an autonomous system. (…) our primary measure (assuming a decent sample size) suggests that the AI and human are performing equivalently. The FDA should be pretty happyª.”)
I think it’s not quite that simple. It’s not true that the FDA doesn’t care about strata, subpopulations, confounders, data subsets/sources/sites, etc. for ML/AI-based devices. For example, the following is from the CADe guidance document on standalone testing (found at https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm187294.pdf):
P. 16: “The study should contain a sufficient number of cases from important cohorts (e.g., subsets defined by clinically relevant confounders, effect modifiers, and concomitant diseases) such that you can obtain standalone performance estimates and confidence intervals for these individual subsets (e.g., performance estimates for different nodule size categories when evaluating a lung CADe device). Powering these subsets for statistical significance is not necessary unless you are making specific subset performance claims”
P. 19: “You should report stratified analysis per relevant confounder or effect modifier as appropriate (e.g., lesion size, lesion type, lesion location, disease stage, imaging or scanning protocols, imaging or data characteristics).”
P. 20: “You should conduct this additional testing in a way that is appropriate and that does not instill additional risk (e.g., increase radiation or contrast exposure) to the patient while still examining the impact of various image acquisition confounders, such as: (…)”
Etc.
There are other relevant FDA documents that consider the issue as well. I find that a good collection of links is https://ncihub.org/groups/eedapstudies/wiki/DeviceAdvice (Device Advice for AI and Machine Learning Algorithms).
Of course that doesn’t cover confounding factors that are unknown or “hidden” which is a problem (but those may not even be part the expert-defined complete schema either, and/or may not be commonly recorded with the image metadata).
LikeLiked by 1 person
Thanks Alex. I agree it was a bit of a simplification, but there are plenty of decision summaries that don’t show application of these rules.
In the pneumothorax example (there is one of these systems approved, no summary online yet) I would assume they show performance by size, but I covering other important variants is less likely.
I’ve heard from people who have gone through the process very variable things, with many saying that this wasn’t done to the level I’m suggesting. If you have a different experience let me know.
The FDA also requires “clinical validation” prior to marketing, but it is the actual interpretation that determines what that means, and most of what they call validation is left to post market monitoring.
LikeLike
Thanks for the response. I agree that the FDA is not perfect 🙂
and that what you are describing needs to be considered seriously by regulatory bodies.
At least, the decision summaries you refer to are probably not for fully autonomous systems, i.e., there is still a human expert in the loop, which lowers the potential risk (I’m actually not sure right now if there are any autonomous AI/ML diagnostic systems other than IDx-DR, though there may be some/many that I’m not aware of).
Regarding “clinical validation”, I think it is relatively well defined at least for radiological CAD devices and is not left for post-market monitoring. For example:
“The clinical performance assessment of CADe devices is typically performed by utilizing a multiple reader multiple case (MRMC) study design, where a set of clinical readers (i.e., clinicians evaluating the radiological images or data in the MRMC study) evaluate image data under multiple reading conditions or modalities (e.g., readers unaided versus readers aided by CADe).” quoted from the FDA guidance document https://www.fda.gov/media/77642/download with a lot more detail on study designs therein as well.
LikeLike
Arguing that they don’t need to care as much because these models are low risk is fine, but then they need to be much more explicit about when they need to start caring more. The have a risk stratification method, but it doesn’t say “at this risk level, stratified testing must be applied” or anything. It sounds quite bespoke at the individual application level.
They also say “Powering these subsets for statistical significance is not
necessary unless you are making specific subset performance claims,” which doesn’t really sound like a recognition that some of these strata might be high-risk and bias the safety profile of the overall task.
And re: clinical validation, all I can say is that the same validation methods applied to mammo CAD, and we know how that turned out. MRMC studies explicitly do not evaluate the safety of a system in an interventional sense.
I could be misunderstanding the FDA position in general, I can only rely on what people have told me about their experiences of the process, and the outcomes we have seen (again, mammo CAD) using the current rules.
Would be nice if they did communicate better, with clear step by step descriptions of the process and more complete summaries of approved product tests (acknowledging the commercial-in-confidence nature of some of it). If anyone from the CDRH wanted to give me the inside scoop, that would be nice 🙂
LikeLike